Predicting chocolate bar ratings
By Alex Labuda
November 26, 2022
In this post, we will be predicting chocolate bar ratings based on a variety of features
The Data
The data includes various chocolate bars and their corresponding ratings
tuesdata <- tidytuesdayR::tt_load(2022, week = 3)
##
## Downloading file 1 of 1: `chocolate.csv`
chocolate <- tuesdata$chocolate
Explore data
First we’ll start by exploring the data
Exploratory data analysis (EDA) is an important part of the modeling process.
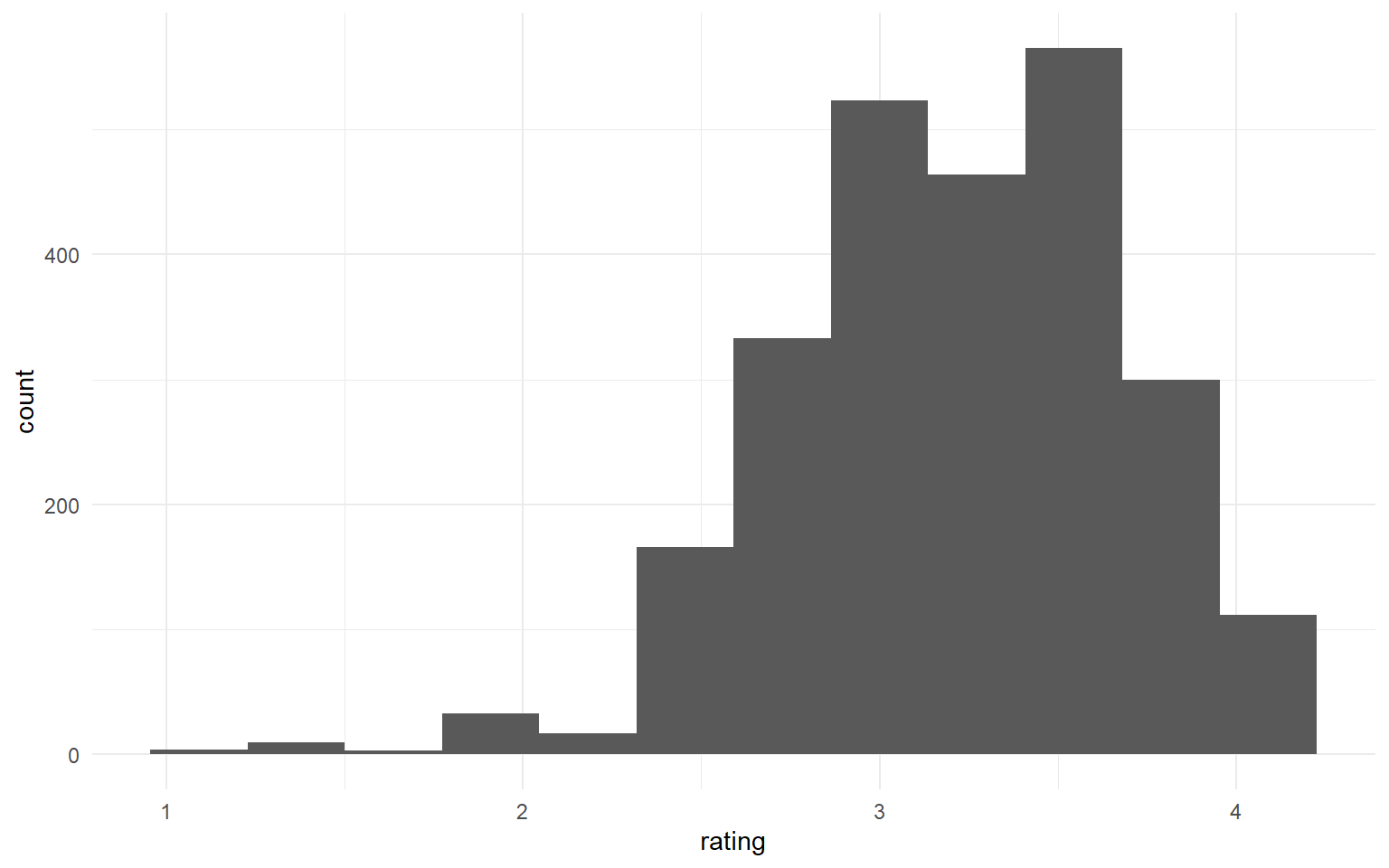
chocolate %>%
ggplot(aes(rating)) +
geom_histogram(bins = 12)
Next I will replace missing values in ingredients with the most common ingredient and remove % from cocoa_percent column
chocolate <-
chocolate %>%
mutate(
ingredients = replace_na(ingredients, "3- B,S,C")
)
chocolate <-
chocolate %>%
mutate(cocoa_percent = as.integer(str_remove_all(cocoa_percent, "%")))
Next I bin all infrequent ingredients, company_location and country_of_bean_origin in “Other” category
chocolate <-
chocolate %>%
mutate(
ingredients = ifelse(!ingredients %in% c("3- B,S,C", "2- B,S"),
"Other",
ingredients),
company_location = ifelse(!company_location %in% c("U.S.A.", "Canada", "France",
"U.K.", "Italy", "Belgium"),
"Other",
company_location),
country_of_bean_origin = ifelse(!country_of_bean_origin %in% c("Venezuela", "Peru",
"Dominican Republic",
"Ecuador", "Madagascar",
"Blend", "Nicaragua"),
"Other",
country_of_bean_origin)
)
Now we can preview out data with skimr::skim()
skimr::skim(chocolate)
Table: Table 1: Data summary
| Name | chocolate |
| Number of rows | 2530 |
| Number of columns | 10 |
| _______________________ | |
| Column type frequency: | |
| character | 6 |
| numeric | 4 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| company_manufacturer | 0 | 1 | 2 | 39 | 0 | 580 | 0 |
| company_location | 0 | 1 | 4 | 7 | 0 | 7 | 0 |
| country_of_bean_origin | 0 | 1 | 4 | 18 | 0 | 8 | 0 |
| specific_bean_origin_or_bar_name | 0 | 1 | 3 | 51 | 0 | 1605 | 0 |
| ingredients | 0 | 1 | 5 | 8 | 0 | 3 | 0 |
| most_memorable_characteristics | 0 | 1 | 3 | 37 | 0 | 2487 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| ref | 0 | 1 | 1429.80 | 757.65 | 5 | 802 | 1454.00 | 2079.0 | 2712 | ▆▇▇▇▇ |
| review_date | 0 | 1 | 2014.37 | 3.97 | 2006 | 2012 | 2015.00 | 2018.0 | 2021 | ▃▅▇▆▅ |
| cocoa_percent | 0 | 1 | 71.64 | 5.62 | 42 | 70 | 70.00 | 74.0 | 100 | ▁▁▇▁▁ |
| rating | 0 | 1 | 3.20 | 0.45 | 1 | 3 | 3.25 | 3.5 | 4 | ▁▁▅▇▇ |
Feature Engineering
Here we will unnest each word in most_memorable_characteristics and examine. We will then use these in our predictions
library(tidytext)
tidy_chocolate <-
chocolate %>%
# puts each word on it's own row
unnest_tokens(word, most_memorable_characteristics)
tidy_chocolate %>%
count(word, sort = TRUE)
## # A tibble: 547 × 2
## word n
## <chr> <int>
## 1 cocoa 419
## 2 sweet 318
## 3 nutty 278
## 4 fruit 273
## 5 roasty 228
## 6 mild 226
## 7 sour 208
## 8 earthy 199
## 9 creamy 189
## 10 intense 178
## # … with 537 more rows
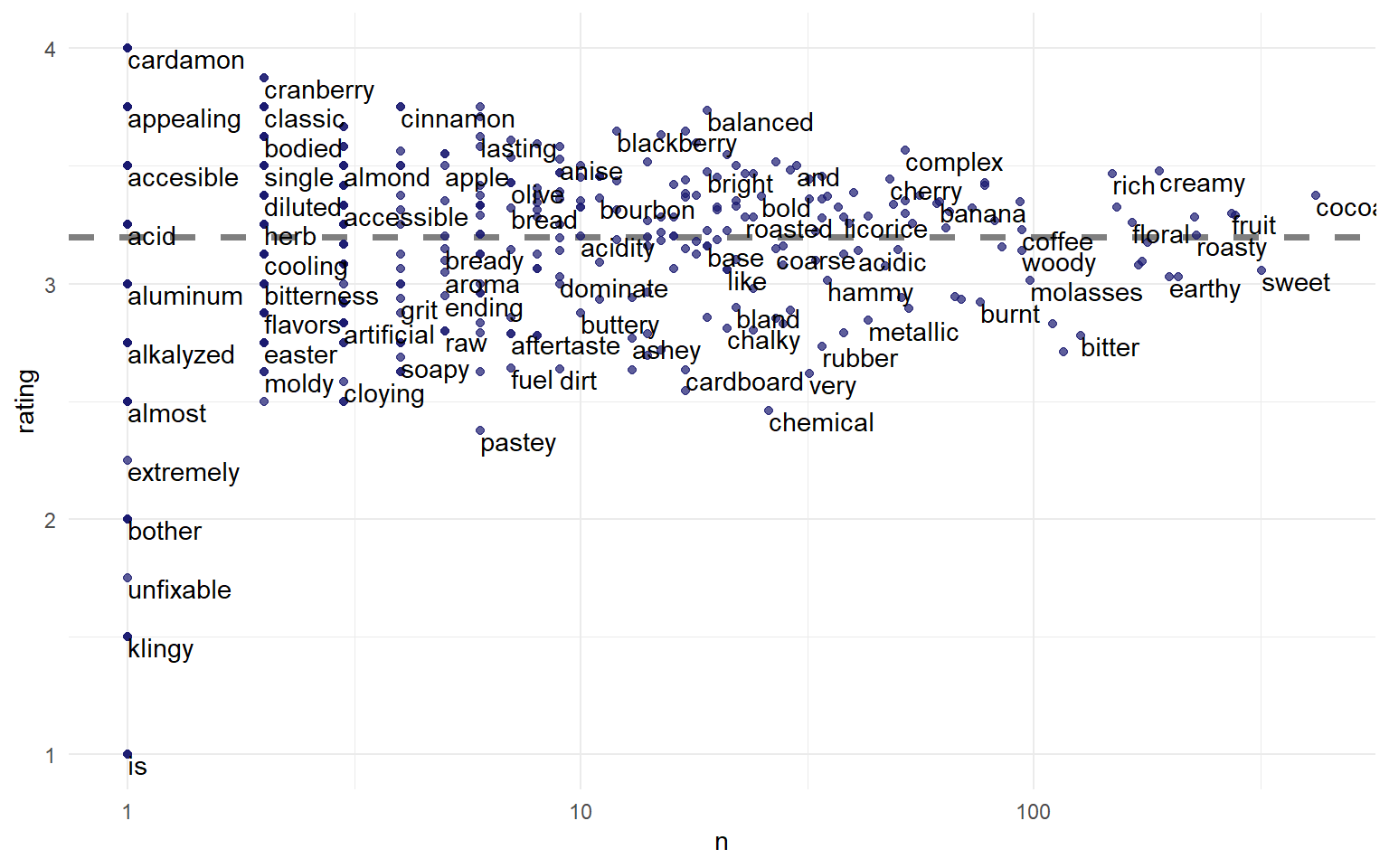
Rating by word count
We can examine most common words in this field
tidy_chocolate %>%
group_by(word) %>%
summarise(n = n(),
rating = mean(rating)) %>%
ggplot(aes(n, rating)) +
geom_hline(yintercept = mean(chocolate$rating),
lty = 2, color = "gray50", size = 1.25) +
geom_point(color = "midnightblue", alpha = 0.7) +
geom_text(aes(label = word),
check_overlap = TRUE, vjust = "top", hjust = "left") +
scale_x_log10()
Build models
Let’s consider how to spend our data budget:
- create training and testing sets
- create resampling folds from the training set
library(tidymodels)
## ── Attaching packages ────────────────────────────────────── tidymodels 1.0.0 ──
## ✔ broom 1.0.1 ✔ rsample 1.1.0
## ✔ dials 1.0.0.9000 ✔ tune 1.0.0
## ✔ infer 1.0.3 ✔ workflows 1.1.0
## ✔ modeldata 1.0.1 ✔ workflowsets 1.0.0
## ✔ parsnip 1.0.2 ✔ yardstick 1.1.0
## ✔ recipes 1.0.1
## ── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
## ✖ scales::discard() masks purrr::discard()
## ✖ dplyr::filter() masks stats::filter()
## ✖ recipes::fixed() masks stringr::fixed()
## ✖ dplyr::lag() masks stats::lag()
## ✖ yardstick::spec() masks readr::spec()
## ✖ recipes::step() masks stats::step()
## • Learn how to get started at https://www.tidymodels.org/start/
set.seed(123)
choco_split <- initial_split(chocolate, strata = rating)
choco_train <- training(choco_split)
choco_test <- testing(choco_split)
set.seed(234)
choco_folds <- vfold_cv(choco_train, strata = rating)
choco_folds
## # 10-fold cross-validation using stratification
## # A tibble: 10 × 2
## splits id
## <list> <chr>
## 1 <split [1705/191]> Fold01
## 2 <split [1705/191]> Fold02
## 3 <split [1705/191]> Fold03
## 4 <split [1706/190]> Fold04
## 5 <split [1706/190]> Fold05
## 6 <split [1706/190]> Fold06
## 7 <split [1707/189]> Fold07
## 8 <split [1707/189]> Fold08
## 9 <split [1708/188]> Fold09
## 10 <split [1709/187]> Fold10
Preprocessing
Let’s set up our preprocessing
Recipe
library(textrecipes)
choco_recipe <-
# recipe for the estimator
recipe(rating ~ most_memorable_characteristics
, data = choco_train) %>%
# steps to build the estimator
step_tokenize(most_memorable_characteristics) %>%
step_tokenfilter(most_memorable_characteristics, max_tokens = 100) %>%
step_tf(most_memorable_characteristics)
# Next steps are not necessary here, but are good to take a look at whats going on
# prep on recipe is analogous with fit on a model
# now the steps say [trained]
prep(choco_recipe)
## Recipe
##
## Inputs:
##
## role #variables
## outcome 1
## predictor 1
##
## Training data contained 1896 data points and no missing data.
##
## Operations:
##
## Tokenization for most_memorable_characteristics [trained]
## Text filtering for most_memorable_characteristics [trained]
## Term frequency with most_memorable_characteristics [trained]
# bake for a recipe is like predict for a model
prep(choco_recipe) %>%
bake(new_data = NULL)
## # A tibble: 1,896 × 101
## rating tf_most_memo…¹ tf_mo…² tf_mo…³ tf_mo…⁴ tf_mo…⁵ tf_mo…⁶ tf_mo…⁷ tf_mo…⁸
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 3 0 0 0 0 0 0 0 0
## 2 2.75 0 0 0 0 0 0 0 0
## 3 3 0 0 0 0 0 0 0 0
## 4 3 0 0 0 0 0 0 0 0
## 5 2.75 0 0 0 0 0 0 0 0
## 6 3 1 0 0 0 0 0 0 0
## 7 2.75 0 0 0 0 0 0 0 0
## 8 2.5 0 0 0 0 0 0 0 0
## 9 2.75 0 0 0 0 0 0 0 0
## 10 3 0 0 0 0 0 0 0 0
## # … with 1,886 more rows, 92 more variables:
## # tf_most_memorable_characteristics_black <dbl>,
## # tf_most_memorable_characteristics_bland <dbl>,
## # tf_most_memorable_characteristics_bold <dbl>,
## # tf_most_memorable_characteristics_bright <dbl>,
## # tf_most_memorable_characteristics_brownie <dbl>,
## # tf_most_memorable_characteristics_burnt <dbl>, …
Specs
Let’s create a model specification for each model we want to try:
ranger_spec <-
# the algorithm
rand_forest(trees = 500) %>%
# engine (computation to fit the model (keras, spark, etc.))
set_engine("ranger") %>% # range is the default
# describes the modeling problem you're working with (regression, classification, etc.)
set_mode("regression")
ranger_spec
## Random Forest Model Specification (regression)
##
## Main Arguments:
## trees = 500
##
## Computational engine: ranger
svm_spec <-
svm_linear() %>%
set_engine("LiblineaR") %>%
set_mode("regression")
svm_spec
## Linear Support Vector Machine Model Specification (regression)
##
## Computational engine: LiblineaR
To set up your modeling code, consider using the parsnip addin or the usemodels package.
Now let’s build a model workflow combining each model specification with a data preprocessor:
WorkFlow
# we're using a workflow bc we have a more complex model preprocess
ranger_wf <- workflow(choco_recipe, ranger_spec)
svm_wf <- workflow(choco_recipe, svm_spec)
If your feature engineering needs are more complex than provided by a formula like sex ~ ., use a
recipe.
Read more about feature engineering with recipes to learn how they work.
Evaluate models
These models have no tuning parameters so we can evaluate them as they are. Learn about tuning hyperparameters here.
doParallel::registerDoParallel()
contrl_preds <- control_resamples(save_pred = TRUE) # keep predictions
svm_rs <- fit_resamples(
svm_wf,
resamples = choco_folds,
control = contrl_preds
)
ranger_rs <- fit_resamples(
ranger_wf,
resamples = choco_folds,
control = contrl_preds
)
How did these two models compare?
collect_metrics(svm_rs)
## # A tibble: 2 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 rmse standard 0.348 10 0.00704 Preprocessor1_Model1
## 2 rsq standard 0.365 10 0.0146 Preprocessor1_Model1
collect_metrics(ranger_rs)
## # A tibble: 2 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 rmse standard 0.344 10 0.00715 Preprocessor1_Model1
## 2 rsq standard 0.379 10 0.0151 Preprocessor1_Model1
We can visualize these results:
bind_rows(
collect_predictions(svm_rs) %>%
mutate(mod = "SVM"),
collect_predictions(ranger_rs) %>%
mutate(mod = "ranger")
) %>%
ggplot(aes(rating, .pred, color = id)) +
geom_abline(lty = 2, color = "gray50", size = 1.2) +
geom_jitter(width = 0.5, alpha = 0.5) +
facet_wrap(vars(mod)) +
coord_fixed()
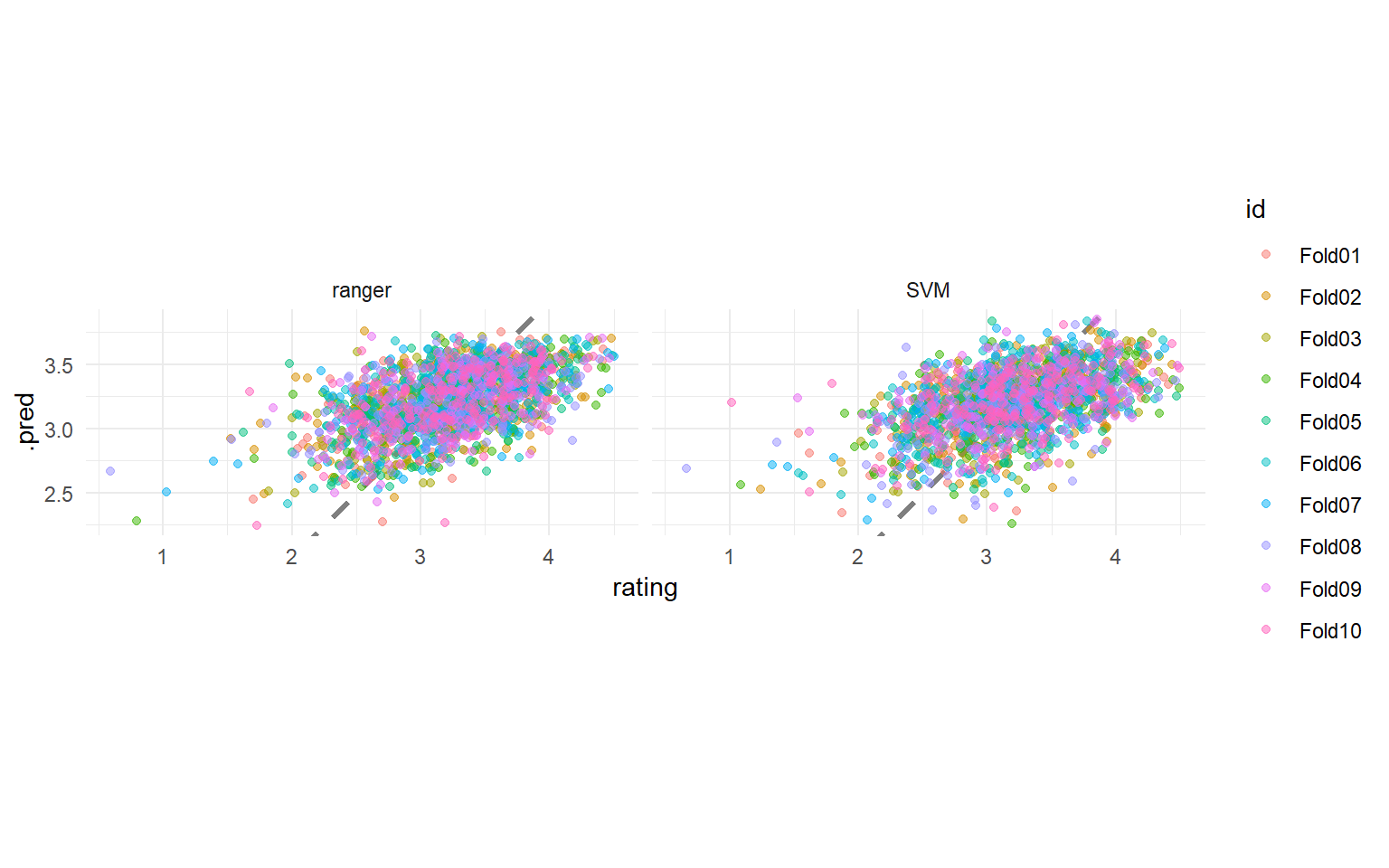
These models perform very similarly, so perhaps we would choose the simpler, linear model. The function last_fit() fits one final time on the training data and evaluates on the testing data. This is the first time we have used the testing data.
final_fitted <- last_fit(svm_wf, choco_split)
collect_metrics(final_fitted) ## metrics evaluated on the *testing* data
## # A tibble: 2 × 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 rmse standard 0.385 Preprocessor1_Model1
## 2 rsq standard 0.340 Preprocessor1_Model1
This object contains a fitted workflow that we can use for prediction.
final_wf <- extract_workflow(final_fitted)
predict(final_wf, choco_test[55,])
## # A tibble: 1 × 1
## .pred
## <dbl>
## 1 3.70
You can save this fitted final_wf object to use later with new data, for example with readr::write_rds().
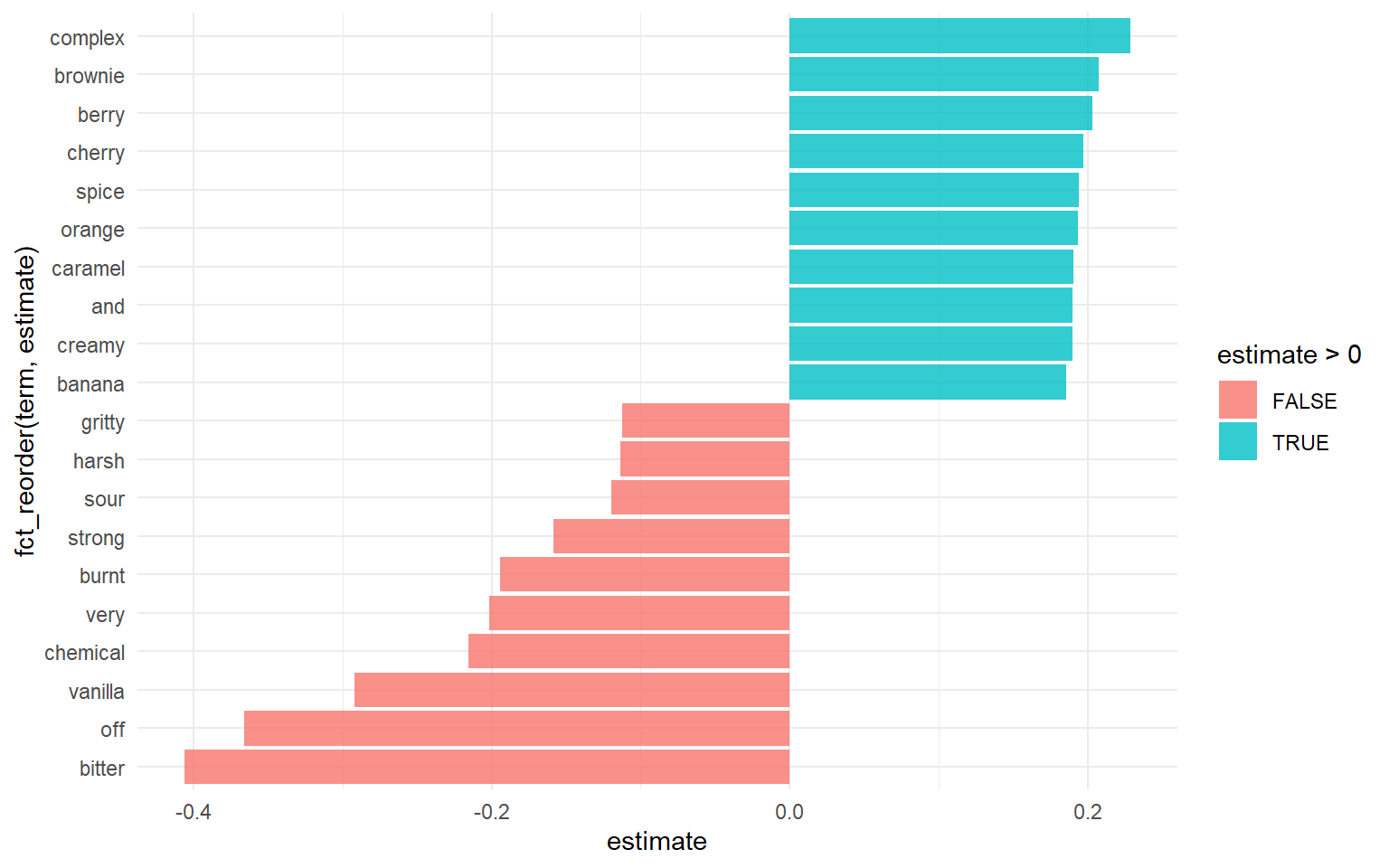
Here we can see words most commonly associated with good ratings, and bad ratings
extract_workflow(final_fitted) %>%
tidy() %>%
filter(term != "Bias") %>%
group_by(estimate > 0) %>%
slice_max(abs(estimate), n = 10) %>%
ungroup() %>%
mutate(term = str_remove(term, "tf_most_memorable_characteristics_")) %>%
ggplot(aes(estimate, fct_reorder(term, estimate), fill = estimate > 0)) +
geom_col(alpha = 0.8)